
Gimias is a software written in C++ for biomedical image computing and simulations. It is equipped with a variety of plug-ins providing technical solutions to specific clinical problems. A number of workflows have been developed in areas including Cardiac Resynchronization Therapy, Aneurysm and Brain Imaging.
Having had no experience with the C++ programming language, I did a self-taught crash course over two weeks, brushing up my skill to a standard where I can handle the more advanced Object-oriented Programming technique used in this specific software. Firstly I integrated a raw code for blood vessels generation from clinical images, into the software as a plug-in with a user interface (in XML) to adjust parameters. I then worked on improving the plug-in for brain vasculature generation to solve a problem where the generated blood vessels intersect and/or fuse with one another (Fig. 1).

Fig. 1. Simulated blood vessels (in blue) overlapped with one another.
A small routine was inserted to revoke newly generated nodes whose radius overlapped with that of a previously generated node. The trick here was that the radius of a daughter vessel is smaller than the parent vessel, ie. the radii of each nodes were compared, instead of simply asserting a fixed value.
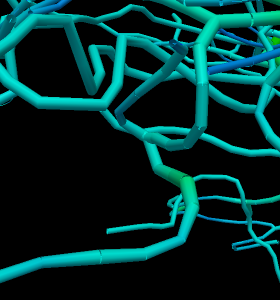
Fig. 2. New algorithms generate vessels which do not overlap or fuse with each other.
The experience in a relatively large research group was eye-opening. It is okay that you work flexible hours, but everyone works very hard to complete their tasks. Defining your own task of the day such that it fits into the bigger picture and leaps a step forward in the long term plan is probably the most important thing to get you going fast. At least that is what I got from this lab.
