Experimentalists: we learnt your effort and understood your hard work.
The last two weeks have been packed with approximately 10 biological experiments, using different techniques. We started with extracting and sequencing our DNA using Sanger sequencing techniques, and drew a phylogenetic tree which proves that, yes I am a Chinese because my mitochondrial DNA shares the highest common features with that in a typical Chinese. We also extracted and sequenced bacterial mutants using Nanopore sequencer. Unfortunately our pair failed to produce any results at the sequencing step possibly due to the traces of ethanol at the end of the drying stage. We took the sequences from successful groups (only a few…) to match with the reference sequences on another sequence analysis software.
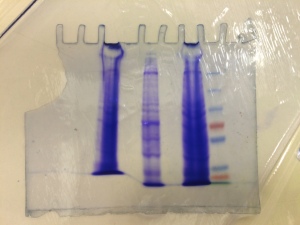
Fig. 1. Our (torn) Coomassie stain: (Left to right) HEK293T (epithelial cell from human embryonic kidney cells), C2C12 (mouse myoblast), N2A (mouse neuroblastoma) and Protein ladder.
Next was Western Blotting – the reason why I chose this course: I have done cell culture with microplate assays before, but never on a Western blot. It turned out that the antibody binding stages had not been done properly (we knew that the trasnfer stage was alright because of the visible bands on Coomassie staining – Fig. 1). There were only faint bands on the X-ray film.
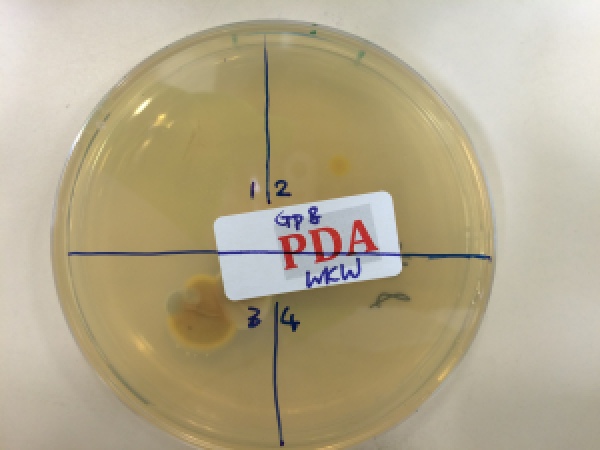
Fig. 2. Potato dextrose agar plate incubated after pressing a finger onto it: (zone 1) With bare fingerprint; (zone 2) With a new sterile glove; (zone 3) With the new sterile glove running through the lab bench; and (zone 4) control.
Whilst waiting for the blots, we did a simple experiment by creating our own “sterile” environment with uplifting airflow (with the aid of a Bunsen burner), only to realise that our lab environment was full of germs – the bacteria grew happily on the agar plates (Fig. 2).
This was followed by two practical of choice:
- Between nematode assay and cockroach electrophysiology: I had been interested in electrophysiology for a while, and thought I would opt for the latter one. We essentially poked the excised cockroach leg with two electrodes to measure the action potential changes when it was simulated. This showed the inherent difficulty in reading extracellular signals accurately and fairly.
- Among confocal, MRI or super-resolution imaging techniques: Consistent with the original idea of taking this course to complement my previous experience, I chose the cardiac MRI practical. The anaesthesia of the mouse had certainly scared the Chemists in my group who had more experience with NMR of single molecules, whilst for me, I found it extremely fascinating how much information we could get from fMRI. This sophisticated my knowledge on clinical imaging modalities and their applications.
With half of the course as Chemists, we studied enzyme kinetics using spectrophotometry – a piece of cake for the Chemists but a little bit of fiddling around for us from the numerical backgrounds.
At the end we grew bacteria with different mutant strains, to study their growth curve using luminescence and qualitative observation of their pathogenicity on mushrooms. Data analysis was the critical point because we would have to understand the possible interpretations of what the mutation did to the bacteria, taking into account the cell growth and functions.
All in all this had been a very useful two-week course for me to broaden my knowledge on wet lab experiments, and perhaps more importantly, understand the tears behind the pretty Western blot images on publications.